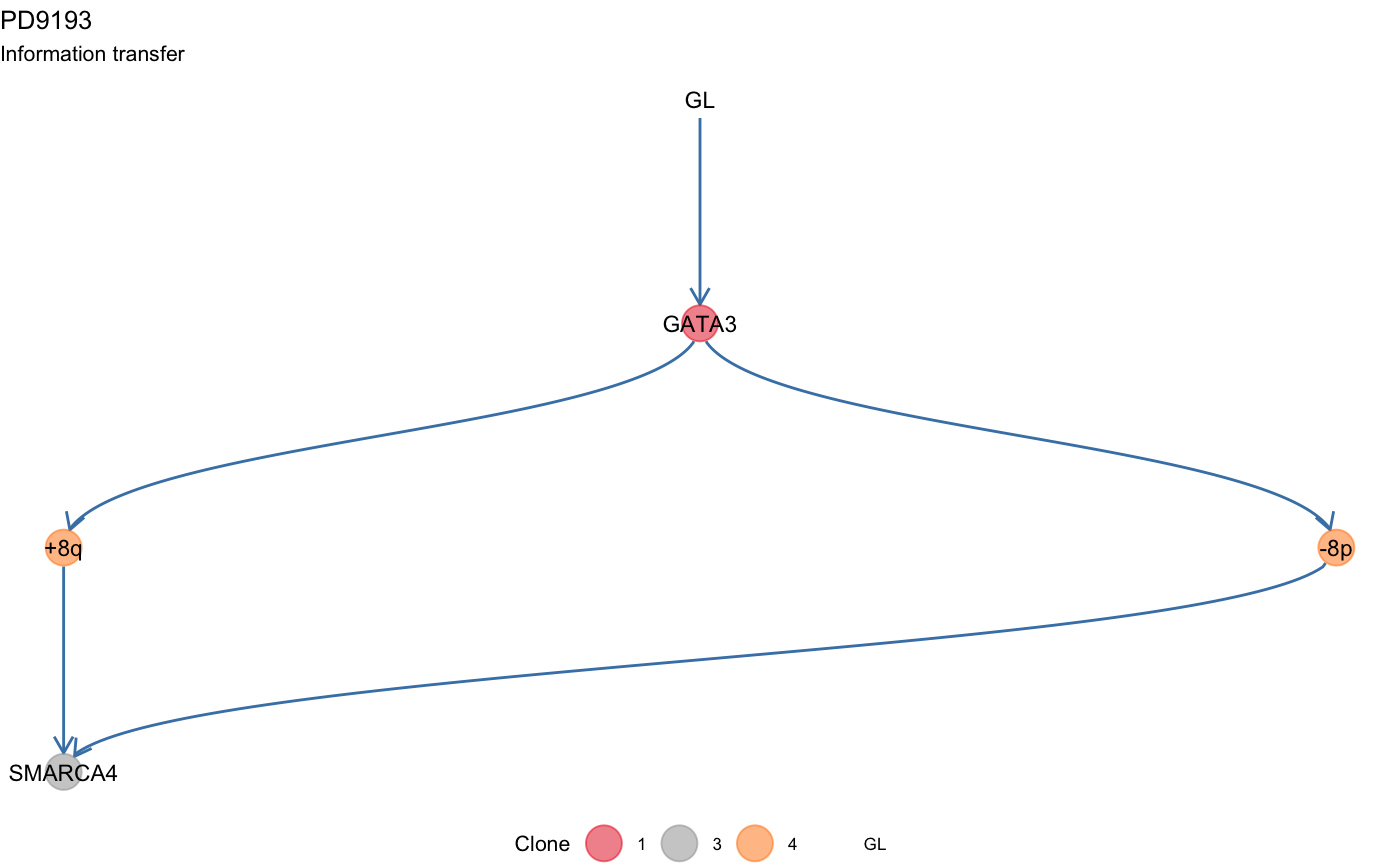
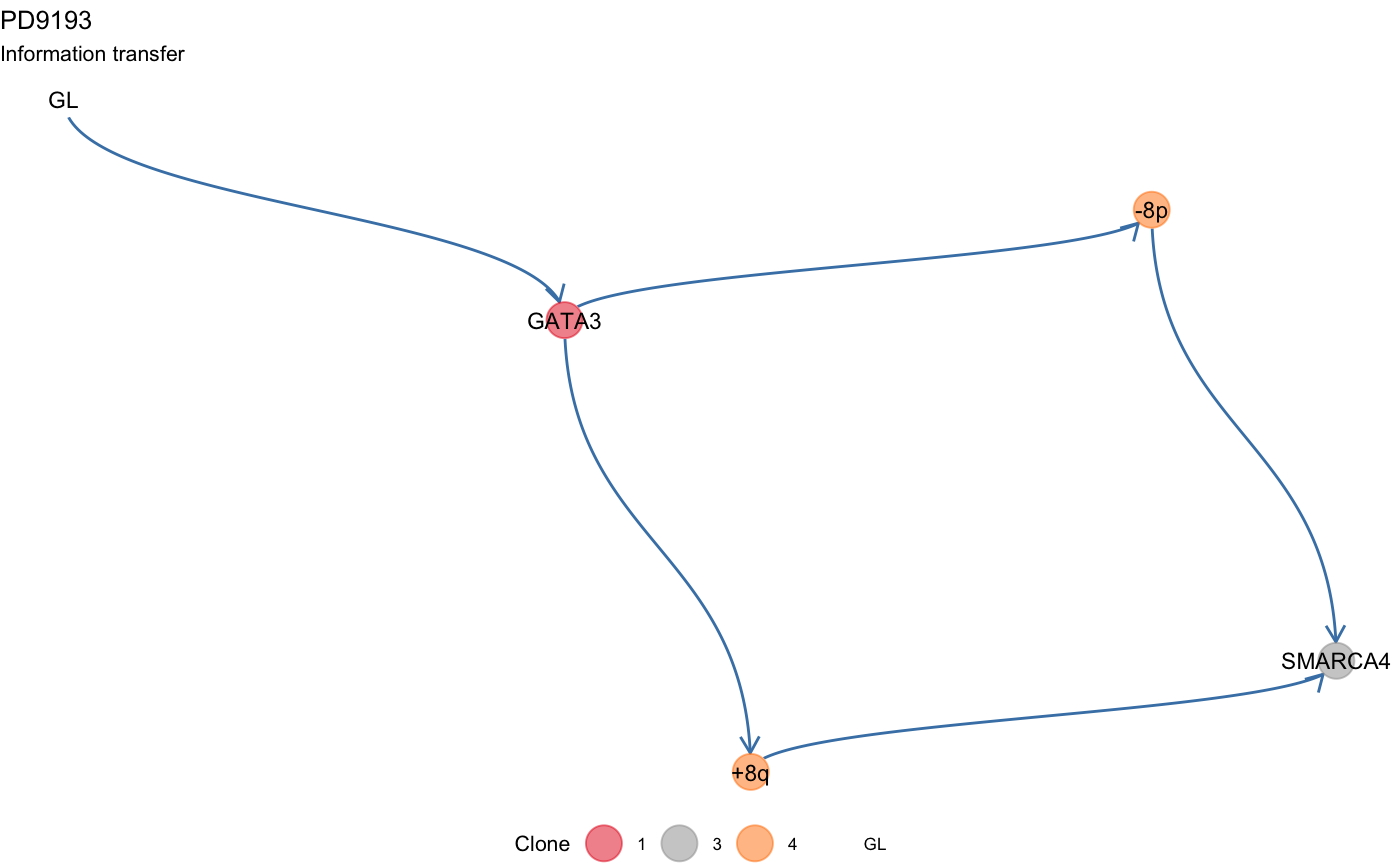
Plot the information transfer of a tree (drivers ordering).
plot_information_transfer.RdThe information transfer of the tree is the set of orderings associated to the internal annotated driver events.
This function plots these orderings following a topological sort of the node of the corresponding clone tree.
plot_information_transfer(x, node_palette = colorRampPalette(RColorBrewer::brewer.pal(n = 9, "Set1")), tree_layout = "tree", ...)
Arguments
| x | A |
|---|---|
| node_palette | A function that can return, for an input number, a number of colours. |
| tree_layout | Layout of this model, as of |
| ... | Other parameters, not used in this case. |
Value
A ggplot object for the plot.
Examples
data(mtree_input) x = mtrees( mtree_input$binary_clusters, mtree_input$drivers, mtree_input$samples, mtree_input$patient, mtree_input$sspace.cutoff, mtree_input$n.sampling, mtree_input$store.max )#> [ mtree ~ generate mutation trees for PD9193 ] #> Sampler : 10000 (cutoff), 5000 (sampling), 100 (max store) #> Suppes' conditions : >= #> # A tibble: 4 x 14 #> Misc patientID cluster is.driver is.clonal PD9193d PD9193e PD9193f PD9193g #> <chr> <chr> <chr> <lgl> <lgl> <dbl> <dbl> <dbl> <dbl> #> 1 cuto… PD9193 2 FALSE FALSE 1 1 1 0 #> 2 cuto… PD9193 3 TRUE FALSE 1 0 0 0 #> 3 cuto… PD9193 1 TRUE TRUE 1 1 1 1 #> 4 cuto… PD9193 4 TRUE FALSE 1 0 1 1 #> # … with 5 more variables: PD9193h <dbl>, PD9193i <dbl>, PD9193j <dbl>, #> # PD9193k <dbl>, nMuts <dbl> #> #> ✔ Structures 3 - search is exahustive #> ✔ Trees with non-zero sscore 3 storing 3 #>plot_information_transfer(x[[1]])# Change layout -- use igraph's "kk" layout plot_information_transfer(x[[1]], tree_layout = 'kk')